
The above picture shows computed ammonia deposition across a 40 by 50 kilometer portion of Europe. It is based on data from 4,000 farms. The map is clear enough, I hope; but its production was a bit of an adventure. As they say, "the devil is in the details." In this article we will explore some of the details, but it will not be necessary to go to hell and back. You can stay right there in front of your computer.
This investigation started as a query on an Internet discussion group (Arcview@listbot.com):
This one has me puzzled. I'm trying to port an emission/deposition model to Arcview/Spatial Analyst. The model itself is very simple and does something like this:
Given a point shapefile containing farm sites (about 5000 farms), we can calculate the emission of Ammonia in kg/yr. The Ammonia emitted from a farm will be deposited in the neighbourhood of the farm. The amount of Ammonia deposited is a function of distance and emission at the farm. The function is available as a table containing distances and deposition factors.
Now, every cell in the resulting deposition grid should contain the sum of all the deposition caused by all the individual farms.
Before we go on, think about how you would solve this problem. In considering this we are going to run into issues that are fundamental to the design of any analytical GIS. In the discussion that follows, ESRI's Spatial Analyst figures prominently as the solution platform, but almost all the design considerations would apply to any raster GIS.
| 1. Convolution |
The first thing is to understand the problem better. Let's consider what goes on in one dimension, where it is easier to draw pictures and see what is happening.
A one-dimensional grid is just a series of equal intervals ("cells") extending from a start point to a later (more positive) end point. The grid has a constant value within each cell.
The value in any cell of our input grid is the total ammonia emitted for all farms within the cell. (If the grid is fine enough, most cell values will be zero, because there will not be farms in every cell. That's ok.) The model calls for ammonia to be distributed among a cell's neighbors in proportion to the emission value in the cell. Here is a picture showing this redistribution (for one input cell only) using arrows:
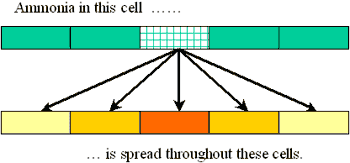
Each arrow represents multiplication of the input cell by a value (that depends on the arrow) and addition of that result to the output cell's value. You have to imagine a similar set of arrows emanating from every input cell.
You can reverse this picture: consider all the arrows arriving at an output cell:
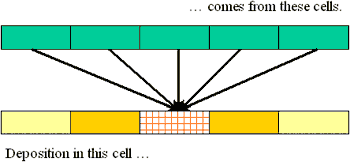
This immediately provides a formula for the value in each output cell, once you provide names for (a) the output cell itself, (b) the input cells, and (c) the multipliers (as represented by the arrows). Conventionally, the arrow factors are known as a "kernel." It is convenient to number the cells left to right, using an arbitrary index such as "i" to designate an arbitrary cell in the grid. Whence
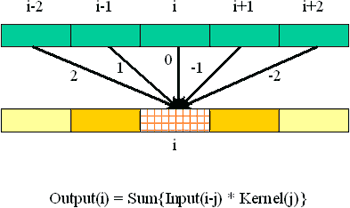
The kernel subscript "j" is an offset relative to the input cell, so that j ranges from +2 down to -2 in our example.
This formula is known as a "convolution." Some grid-based GISes offer it as a built-in capability, where of course it has been programmed to convolve any two arbitrary grids efficiently. It is tempting to treat this capability as a "black box" and just use it, but as you will see-keep reading!-there is much to be gained by understanding its innards.
This formula for the convolution is really output oriented, or neighbor-centric: it explicitly describes how each of cell i's neighbors (i-j) contributes to the deposition there. This leads to the obvious algorithm of computing the convolution cell by cell:
For each output cell i {
Set Output(i) to 0.0
For each kernel cell j {
Add Kernel(j) * Input(i-j) to Output(i)
}
}
Algorithm 1: Convolution
The number of numerical operations required for this computation evidently is proportional to the number of output cells times the number of kernel cells. This is true regardless of the dimension of the grid.
| 2. Analysis of computing requirements |
To learn more, I got in touch with Marc Hoogerwerf, who graciously provided more documentation. The farms are in a densely populated part of Europe and often are situated less than 100 meters apart, so his grids have cells smaller than 100 meters. The ammonia emissions can spread as much as three kilometers-at least 30 cells-away from each farm in all directions, although they taper off quickly after the first several hundred meters. His immediate study area is almost 40 by 50 kilometers, containing about 4,000 farms with emissions data. (Some of the original 5,000 farms presently have no data.)
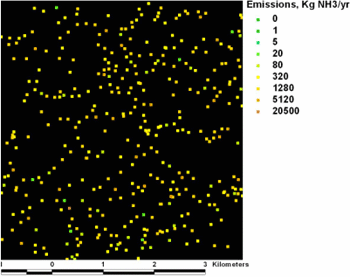
A small portion of the input grid. What do you think the deposition pattern might look like?
Spatial Analyst offers some convolution-like capabilities. Its "FocalStats" request, for example, can compute a local moving-window average. It is not hard to see that this is a convolution (think about how it would work on a one-dimensional grid). The kernel is the size of the moving window and all its values are equal to 1/N, the reciprocal of the number of cells in the kernel. By playing some map algebra tricks, FocalStats can be persuaded to yield a convolution (more about that later).
Hoping to press FocalStats into service, I measured some computation times on large random grids, systematically varying the kernel size from 1 x 1 through 20 x 20. (Spatial Analyst cannot compute with kernels larger than 20 x 20.) The times varied as expected from Algorithm 1. Here are some of the predicted times, in seconds, to do the computation on a 512 x 512 grid on my computer, a 200 MHz Pentium Pro:
Predicted computation times for moving averages.
| Kernel size | Time in seconds |
| 1 x 1 | 0.09 |
| 2 x 2 | 0.38 |
| 4 x 4 | 1.50 |
| 6 x 6 | 3.38 |
| 8 x 8 | 6.01 |
| 12 x 12 | 13.53 |
| 20 x 20 | 37.58 |
These values have been adjusted to reflect the processing time used after the input grid has been read but before the output is written to disk. They are consistent with performing about 2.8 million operations (of one multiplication and one addition) per second.
Using 25-meter cells, we will need a grid of about 1600 by 2000 cells for the input (and the output) and 240 by 240 cells (covering a circle of radius three kilometers) for the kernel. Using this gridded representation of the data, the total number of arithmetic operations will be proportional to the product of the output and kernel grid sizes, or (1600 * 2000) * (240 * 240) = 200,000 million operations, more or less. Let's see-that's about 70,000 seconds per FocalStats request, and we might need a lot of those requests to do one full convolution.
Exercise: How many days of computation is this?
The origin of this project.
Here is Part Two.
Here is Part Three.